
[ad_1]
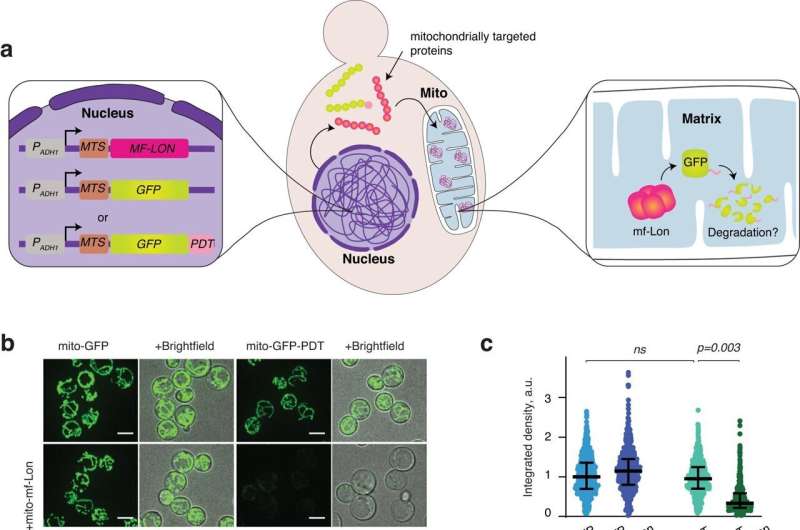
Mitochondria-specific degradation of GFP-PDT by M. florum Lone protease. A schematics showing the experimental setup. b Representative live cell images of yeast cells expressing mitochondrially targeted GFP or GFP-PDT, either independently (upper panels) or together with mitochondrially targeted M. florum Lone (lower panels). c Dot plots show quantification of GFP intensity as arbitrary units (au) of the indicated yeast strains, each dot representing an individual cell. Similar to db , but strain with GFP lacking MTS is shown. As in ec, except that the dot plots show the GFP intensity of cells in d, and the data are normalized to the cyto-GFP strain. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-45819-6
A new study published I Nature Communications Karolinska Institutet researchers have solved a long-standing problem by establishing a system that allows site-specific protein depletion within mitochondria, the cellular centers of energy production and metabolism.
Understanding how cells work often requires manipulation. Protein function. The methods used to do this usually result in complete loss of protein function and cannot provide information about their specific roles in different cellular compartments. This is particularly difficult for organelles such as mitochondria.
To address this, the researchers presented, for the first time, a technique for targeted protein degradation within yeast mitochondria. Human cells. They also devised a method to control the inclusion of decay, thus allowing time-resolved analysis.
An important aspect of this technique is the ability to isolate mitochondrial function from the rest of the cell. Being of bacterial origin, mitochondria have a certain degree of autonomy, such as their own genome, a complex system. Energy productionand replicability.
Nevertheless, proper mitochondrial function is highly dependent on other organelles, including the nucleus. When crosstalk between mitochondria and other organelles fails, mitochondrial dysfunction Consequently, which is strongly correlated Neurodegenerative diseases and age-related diseases such as cancer.
Not surprisingly, specific targeting of mitochondrial function is therefore essential to understand and eventually treat such disorders.
“We introduced a bacterial protease into yeast and human mitochondria to achieve a mitochondrial-specific protein deficiency. We did this with the knowledge that mitochondria are evolutionarily derived from bacteria, and a bacterial protease This is how mitochondria must function in the environment.
“We chose to use a lone protease from a mollicute, which detects and degrades a protein tag called PDT. It can be added to the protein under investigation by genetic engineering.
“To our great excitement, we found that Lon protease was able to degrade PDT-tagged proteins specifically within mitochondria in yeast and human cells,” says Camila Bjorkgren, professor in the Department of Cell and Molecular Biology.
Swastika Sanyal, who developed the system, says there are plans to take the project further. “Our next step will be to use the PDT tagging system we developed to identify the mitochondria-specific functions of DNA-maintaining enzymes. Many of these proteins also bind to nuclear DNA. maintain, so by reducing them only within the mitochondria, we can identify their direct role in mtDNA repair.
“This study will enable us to better understand what goes wrong during mitochondrial dysfunction, which can result from altered and deleted mtDNA.”
More information:
Swastika Sanyal et al., A system for inducible mitochondria-specific protein degradation in vivo. Nature Communications (2024). DOI: 10.1038/s41467-024-45819-6
Provided by
Karolinska Institute
Reference: New technique developed for targeted protein degradation
This document is subject to copyright. No part may be reproduced without written permission, except for any fair dealing for the purpose of private study or research. The content is provided for informational purposes only.
[ad_2]


