
[ad_1]
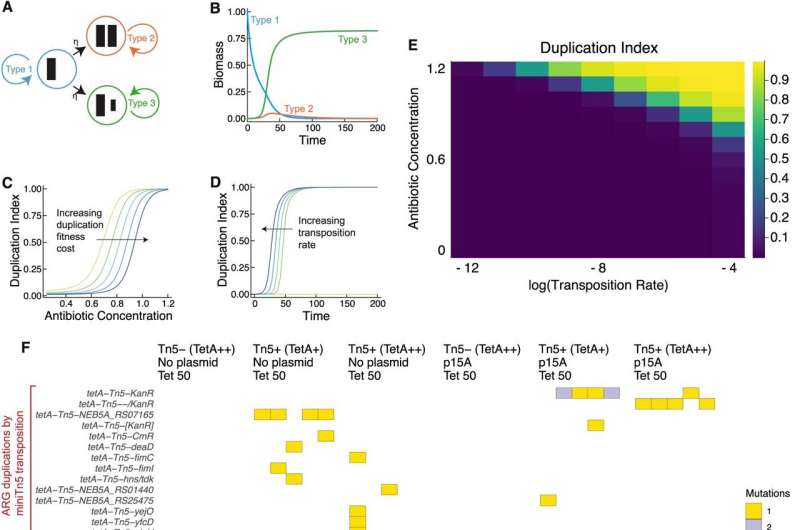
Mathematical modeling and laboratory evolution with E. coli K-12 DH5α demonstrate that antibiotic selection is sufficient to drive the rapid evolution of antibiotic resistance through antibiotic resistance gene transcription. Credit: Nature Communications (2024). DOI: 10.1038/s41467-024-45638-9
Duke University biomedical engineers have uncovered an important link between the spread of antibiotic resistance genes and the evolution of resistance to new drugs in certain pathogens.
Research shows. Bacteria Exposure to high levels of antibiotics often results in multiple identical copies of protective antibiotic resistance genes. These duplicated resistance genes are often linked to “jumping genes” called transposons that can jump from strain to strain. Not only does this provide a mechanism for resistance to spread, but having multiple copies of a resistance gene can also provide a handle for evolution to develop resistance to new types of drugs.
Results to become visible In the journal Nature Communications.
Early work from the Lingchong Yu lab showed that 25 percent of bacterial pathogens have the ability to spread antibiotic resistance through horizontal gene transfer. They have also shown that the presence of antibiotics does not accelerate the rate. Horizontal gene transferso there is something else going on that forces the genes to spread.
“Bacteria are constantly evolving under many pressures, and the increased transcription of certain genes is like a fingerprint. Event This allows us to see what kinds of functions are evolving really quickly,” said James L. Merriam Distinguished Professor of Biomedical Engineering at Duke, a postdoctoral fellow working in Lingchong Yu’s laboratory. “We hypothesized that bacteria are invaders. Antibiotics often have multiple copies of protective resistance genes, but until recently we didn’t have the technology to find the smoking gun.”
Traditional DNA-reading technology copies short fragments of genes and counts them, making it difficult to determine whether high concentrations of specific sequences are actually present in a sample or whether the reading process has artificially detected them. is being extended. However, in the past five years, whole-genome sequencing with long-read technology has become more common, allowing researchers to detect high levels of genetic duplication.
In the study, Maddamsetti and co-authors quantified the frequency of resistance genes present in samples of bacterial pathogens taken from a variety of environments. They discovered that people living in places with high antibiotic use—humans and livestock—were enriched with multiple identical copies of antibiotic resistance genes, while wild plants, animals, soil, and water. Such transcripts are rare in living bacteria.
“Most bacteria have some basic antibiotic resistance genes, but we’ve rarely seen them replicate in nature,” you said. “In contrast, we’ve seen a lot of replication in humans and cattle where we’re potentially hammering them with antibiotics.”
The researchers also found that levels of resistant transcripts were even higher in samples taken from clinical datasets where patients were likely taking antibiotics. This is an important point, he says, because antibiotic copying increases Resistance genes It also increases the chances of bacteria developing resistance to new types of treatment.
“Making continuous copies of genes for resistance to penicillin, for example, could be the first step towards being able to break down a new type of drug,” said Madam Setti. “It gives evolution more rolls of the dice to find a particular mutation.”
“Everyone recognizes that the antibiotic resistance crisis is growing, and the knee-jerk reaction is to develop new antibiotics,” you added. “But what we find over and over again is that if we can figure out how to use antibiotics more efficiently and effectively, we can potentially solve this crisis by more than just developing new drugs. can be effectively resolved.”
“Most antibiotics used in the United States are not used on patients, they are used in agriculture,” you added. “So this is a particularly important message for the livestock industry, which is one of the main reasons why. Antibiotic resistance Always out and getting more serious.”
More information:
Rohan Maddamsetti et al., Duplicated antibiotic resistance genes reveal ongoing selection and horizontal gene transfer in bacteria, Nature Communications (2024). DOI: 10.1038/s41467-024-45638-9
Provided by
Duke University
Reference: Study Finds a Smoking Gun for the Spread and Evolution of Antibiotic Resistance (2024, February 22) Accessed February 23, 2024 at https://phys.org/news/2024-02-gun-evolution-antibiotic-resistance Retrieved from .html
This document is subject to copyright. No part may be reproduced without written permission, except for any fair dealing for the purpose of private study or research. The content is provided for informational purposes only.
[ad_2]


