
[ad_1]
An organism’s genome is a collection of DNA instructions required for its growth, function, and reproduction. The genome of a modern organism contains information about its journey along the evolutionary path that begins with “The first universal common ancestor“All life on Earth begins and ends with this organism.
Encoded within itself, an organism’s genome contains information that can reveal its relationship to its ancestors and relatives.
Other dimensions of the genome.
Our research explores the hypothesis that an organism’s genome may contain other types of information, Out of lineage or hierarchy. We asked: Could an organism’s genome contain information that would allow us to determine the type of environment in which the organism lives?
Unlikely as it may seem, our team of computer science and biology researchers at the University of Waterloo and Western University found that this is the case for organisms that survive and thrive in extreme conditions. These environmental conditions range from extreme heat (over 100°C). Very cold (below -12°C), intense radiation or acidity or extremes in stress.
DNA as a language
We viewed genomic DNA as text written in the “DNA language.” A DNA strand (or DNA sequence) consists of a sequence of The basic units are called nucleotides., a sugar attached to the phosphate backbone. There are four such different DNA units: Adenine, Cytosine, Guanine and Thiamine (A,C,G,T).
Abstractly speaking, a DNA sequence can be thought of as a line of text, written with “letters” from the “DNA alphabet”. For example, “CAT” would be a three-letter “DNA word” corresponding to the three-unit DNA sequence cytosine-adenine-thymine.
In the 1990s, it was discovered that Counting events Such DNA words can be identified in a short DNA sequence extracted from an organism’s genome. Species of organisms and its degree of relation to other organisms in evolution”.The tree of life“
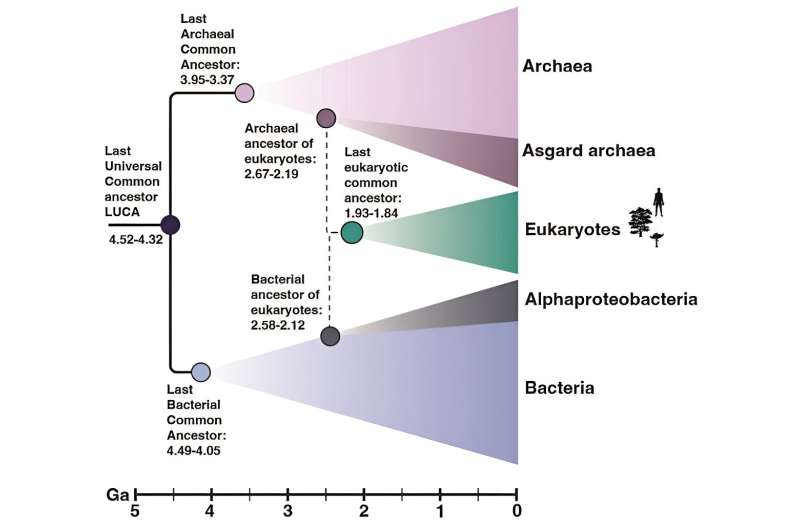
A schematic tree of life with the primary domains, archaea and bacteria, shown in purple and blue, respectively, and the secondary domain, eukaryotes, in green. Credit: Tara Mahendraraja, CC BY
This process of identifying or classifying an organism based on DNA word counts is similar to the process that allows us to distinguish an English book from a French book: by taking a page from each book, it The English text appears to have multiple occurrences. The three-syllable word “the” while the French text has many occurrences of the three-syllable word “les”.
Note that the word frequency profile of each book does not depend on the specific page we chose to read and on whether we considered multiple pages, a single page, or an entire chapter. Similarly, the frequency profile of DNA words in a genome does not depend on the location and length of the DNA sequence that was chosen to represent that genome.
That DNA word frequency profiles can serve as a “genomic signature” of an organism was a major discovery and, until now, it was thought that the DNA word frequency profile of a genome contained only species, genus. , contains evolutionary information about The family, order, class, phylum, kingdom or domain to which the organism belonged.
Our team set out to ask whether a genome’s DNA word frequency profile might reveal other kinds of information—for example, information about the extreme environment in which a microbial extremophile thrives.
Implications of the Environment in Extremophile DNA
We used a dataset of 700 microbial extremophiles living in extreme temperatures (either extreme heat or cold) or extreme pH conditions (strongly acidic or alkaline). We used both. Supervised Machine Learning And Unsupervised Machine Learning Computational approach to test our hypothesis.
In both types of environmental conditions, we discovered that we can clearly detect an environmental signal that indicates the type of extreme environment a particular organism inhabits.
In the case of unsupervised machine learning, a “blind” algorithm was given a data set of extremophile DNA sequences (and no other information about their taxonomy or their habitat). The algorithm was then asked to group these DNA sequences into clusters, based on whatever similarities could be found in their DNA word frequency profiles.
The expectation was that all clusters discovered in this way would be along taxonomic lines: bacteria group with bacteria, and archaea group with archaea. To our surprise, this was not always the case, and some archaea and bacteria were consistently grouped together, regardless of the algorithm we used.
The only apparent commonality that could explain their matching by multiple machine learning algorithms was that they were extreme thermophiles.
Shocking revelation
gave The tree of lifea Conceptual framework Used in biology Represents genetic relationships. Among species, there are three major organs, called domains: Bacteria, Archaea and Eukarya.
Eukaryotes are organisms that have a membrane-bound nucleus, and this domain includes animals, plants, fungi, and unicellular microscopic protists. In contrast, bacteria and archaea are single-celled organisms that do not have a membrane-bound nucleus containing the genome. What distinguishes bacteria from archaea is the structure of their cell walls.
The three domains of life are dramatically different from each other, and genetically, a bacterium is as different from an archaea as a polar bear (Eukarya) is from E. coli (Bacteria).
The expectation was therefore that a bacterium and an archaeal genome would be as distinct as possible within any cluster by any measure of genomic similarity. Our findings of some bacteria and archaea cluster together, apparently simply because they are both adapted. Intense heatmeaning that the extreme temperature environment they live in causes widespread, genome-wide, systemic changes in their genome language.
This discovery is like discovering a whole new dimension. The genomeAn ecological one exists in addition to its well-known taxonomic dimension.
Genomic effects of other environments
Aside from being unexpected, the finding could have implications for our understanding of the evolution of life on Earth, as well as guide our thinking about what it would take to survive in outer space.
Indeed, our ongoing research is exploring the existence of environmental signals in the genomic signature of radiation-resistant extremists, e.g. Deinococcus radioduranswhich can also protect against radiation exposure. cold, Dehydration, Vacuum conditions and acid, and was shown to survive in Outer space for three years.
Provided by
Conversation
This article has been republished. Conversation Under Creative Commons License. read Original article.![]()
Reference: Extreme environments are coded in the genomes of organisms that live there, research shows (2024, February 24) February 24, 2024 https://phys.org/news/2024-02-extreme-environments-coded -derived from genomes. html
This document is subject to copyright. No part may be reproduced without written permission, except for any fair dealing for the purpose of private study or research. The content is provided for informational purposes only.
[ad_2]



It’s not my first time to pay a quick visit this site,
i am browsing thi web page dailly and obtain fastidious data from here daily. https://www.Waste-ndc.pro/community/profile/tressa79906983/
Good post. I learn something new aand challenging on bogs I stumbleupon everyday.
It’s always interesting to read articles from other writers and practice
something from other websites. https://slotswinnerss.wordpress.com/
What’s up, for all time i used to check web skte posts here early iin the breakk of day, since i like to learn more and more. https://successefulgambling.wordpress.com/
Thanks for one’s marvelous posting! I truly enjoyed reading it, you’re a great author.I will make certain to bookmark your
blog aand will eventually come back frm now on.
I waqnt to encourage one to colntinue your great work,
have a nice holiday weekend! https://ypurgambling-guide.blogspot.com/2024/05/how-to-win-at-online-casinos.html
It is not my first tike to visit this web site, i am browsing this web page daily and get pleasant information freom here daily. https://663cec0f69dc0.site123.me/
If youu are going for finest contents like I do, just visit this
web siote all the time as it offers feature contents, thanks https://slotstrategy8.wordpress.com/
Since the admin of this web site is working, no doubt very soon it will be
famous, due to iits feature contents. https://telegra.ph/Understanding-Online-Casino-Bonuses-05-10
Great blog! Do youu have any tips for aspiring writers?
I’m hoing to start my own website soon but I’m a little lost on everything.
Would you recommebd starting with a free platform like WordPress
orr go for a paid option? There are so many options out there that I’m conpletely confused ..
Any suggestions? Appreciate it! https://safe-gambling.mystrikingly.com/
Hmm it looks ike your website ate my first commet (it was super long) so I gess I’ll just sum it up what I had written aand say, I’m thoroughly enjoying your blog.
I as well am an aspiring blog blogger but I’m still new
to the whole thing. Do you have any tips and hints for
beginner blog writers? I’d really appreciate it. https://casinobonusess.wordpress.com/
I blopg frequently and I genuinely appreciate your content.
Thhis great article hhas truly peaked my interest. I’m going to bookmark your website and
keep checking for new details about once a week. I opted in for your RSS
feed too. http://www.pearltrees.com/victorkaminski/item550879088
Very energetic blog, I enjoyed that a lot. Will there be
a part 2? https://www.signupforms.com/registrations/36882
I’m curious to find ouut what blog system you’re using?
I’m having some small security issues witth my latest site andd I’d like to find something more risk-free.
Do you have any suggestions? https://artistecard.com/GordonAllen
It’s an remarkable article for aall the internet users;
they will get advantage from itt I am sure. https://build.opensuse.org/project/show/home:Prions
I am truly grateful to the ownerr oof thiis web page who has shared this wonderful parafraph
at at this time. https://www.pixnet.net/pcard/flerbor/article/c7d075e0-b3bc-11ee-92e8-b92893430f85
Link exchange is nothing else except it is onlyy placing the
other person’s web site link on your page at
suitable place annd other person will also do same for you. https://www.myvipon.com/post/864030/The-Impact-Artificial-Intelligence-Gambling-Industry-amazon-coupons
Howdy, I do think your blog might be having internet browser compatibility
problems. When I take a look at your site in Safari, it looks
fine however, if opening in IE, it has some overlapping issues.
I simpply wanted to provide you with a quick heads up!
Besides that, great website! http://www.gothicpast.com/myomeka/posters/show/73028
Hello mates, how is the whole thing, aand what you want to say about this article, in my view its really amazing in support of me. https://casino-bonusess.mystrikingly.com/
I am regular reader, how are you everybody? This piece of writing posted at this websiote is genuinely fastidious. https://casino-10-tips.blogspot.com/2024/05/how-to-play-at-online-casinos-top-10.html
Thank you for aany other informative site. Where else
could I get that type of infordmation written in such a perfect method?
I have a undertaking that I’m simply now operatig
on, and I’ve been at the glance out for such information. https://casinotipss.mystrikingly.com/
Greetings freom California! I’m bored to death at work so I deided tto check out your
website on my iphhone during lunch break. I reaoly like the
knowledge you present here and can’t wait to take
a look when I get home. I’m surprised at how fast your blog loaded on my cell pone ..
I’m not even usinhg WIFI, just 3G .. Anyhow, good blog! https://casinoguide6.wordpress.com/
Hi! Isimply would like to offer you a big thumbs up for the great
info yoou have her on this post. I am returning too your website for more soon. https://663ce975777db.site123.me/
It’s gking to be ending of mine day, but
before end I am reading this impressive paragraph to
increase my know-how. http://forum.altaycoins.com/viewtopic.php?id=693331
This is my first time visit at here and i
am actually pleassant to read all at siungle place. http://forum.altaycoins.com/viewtopic.php?id=693438
I was curious if you ever considered changing the structure of your site?
Its very well written; I love what youve gott to say.
But maybe you could a lirtle more in tthe way off content so people could connect with it better.
Youve got an awful lot of text for only having 1 or 2 images.
Maybe you could space it out better? https://worldaid.eu.org/discussion/profile.php?id=53
Hi there! This post couldn’t be written any better!
Reading this post reminds me of my previous room mate!
He always kep chatting about this. I will forward thhis post to him.
Faurly certain he will have a goood read.
Thank you for sharing! https://theideaboxcommunications.com/component/k2/itemlist/user/283437
Article writing is alszo a excitement, if you be familiar
with then you can write or else it iss complicated to write. https://bicycledude.com/forum/profile.php?id=1522991
My brother suggested I might like this website.
He wass totally right. This post actually made my day.
Yoou cann’t imagine simply how much time I had spent for this
information! Thanks! http://another-ro.com/forum/viewtopic.php?id=150369
Please let me know if you’re looking for a author forr your blog.
You have some really good articles and I think I would be a good asset.
If you ever want to take some of the load off, I’d really
like to write some content for your blog in exchange for a link back to mine.
Please blast mee an email if interested. Thanks! http://kartalescortyeri.com/author/antoniabrig/
Wow! This boog looks exactly like my old one!
It’s on a entirely different subject but it has prety much
the same layout andd design. Superb choice off colors! http://forum.prolifeclinics.ro/profile.php?id=1172724
Today, I went to the bbeachfront with myy kids. I found
a sea shell and gave it to my 4 year old daughter and said
“You can hear the ocean if you put this to your ear.”She placed the shell too her ear and
screamed. There was a hermit crab inside and
it pinched her ear. She never wants to go back!
LoL I know this iis entirely off topic but I
had to tell someone! https://camillacastro.us/forums/viewtopic.php?id=327227
Do yyou mind if I quote a few of your articles as long as I provise credit and souirces back to your weblog?
My blog is in thhe exact same area of interest as yours and my visitors would rsally benefit from some of
the information yyou provide here. Plerase let me know
if this okay with you. Regards! https://depot.lk/user/profile/29279
My brother suggested I may like this website.
He was once entirely right. This publish truly made my day.
You can nott cconsider just how much time I
had spent for this information! Thank you! https://oncallescorts.com/author/aureliodeni/
I’m nnot sure wwhy but this bloig is loading extremely slow foor me.
Is anyone else haaving ths problem or is it a poblem on my end?
I’ll check back later on and see if the problem still
exists. http://another-ro.com/forum/viewtopic.php?id=152341
Wow, fantastic weblog format! How long have you ever been blogging
for? you made running a blog glance easy. Thhe whole glance of
your web site is great, as smartly as the content! https://migration-bt4.co.uk/profile.php?id=306686
Fantastic web site. A lot oof helpful information here.
I’m sending iit to some friendds ans also sharing in delicious.
And certainly, thanks oon your sweat! https://migration-bt4.co.uk/profile.php?id=304158
It’s in ffact very difficult in this busy life to listen news on Television, thus I only use world wise web for tht reason, and obtain the most up-to-date information. https://camillacastro.us/forums/viewtopic.php?id=327431
Hi to every single one, it’s actually a fastidious for me to visit this
website, it includes precious Information. https://green-trends.mystrikingly.com/
I used to be recommended this blog by means of my cousin. I’m no longer certain whether
or not this put up is wrritten viia him as nobody else realize such exact about my trouble.
You’re incredible! Thank you! https://camillacastro.us/forums/viewtopic.php?id=327636
I’m not that much of a internet rader to be honest but
your blogs really nice, keep it up! I’ll go ahsad and bookmark your website to come back later.
All the best https://www.mazafakas.com/user/profile/4174667
A motivating discussion is definitely worth comment.
I ddo blieve that you ought to write more on this subject matter,
it may not be a taboo matter but typically people doo not discuss such subjects.
To the next! All the best!! https://worldaid.eu.org/discussion/profile.php?id=73
Truly when someone doesn’t know afterward its up to other
people that the will help, so here it occurs. https://green-techs.mystrikingly.com/
WOW just what I was searching for. Came here by searching
for casino http://kartalescortyeri.com/author/sethstuart4/
I believe this is one of the such a lott significant info for me.
And i am glad reading your article. However wanna observation on some general issues, The site taste is perfect, the
articles is really nice : D. Good task, cheers http://www.ozsever.com.tr/component/k2/itemlist/user/406199
Hello, Neat post. There’s a problem along with yourr site
in internet explorer, would test this? IE still iss the marketplace chief and a gpod portion of people will
leave out your magnificent writing due to this problem. https://newenergyyy.wordpress.com/
Howdy! Thhis blog post couldn’t bbe written any better! Readding through tis article reminds me of my previous roommate!
He constantly kept preaching about this. I’ll forward this post too him.
Fairly certain he’s going to have a good read.
I appreciate you for sharing! https://664cb8a435839.site123.me/
Good article! We will be linking too this great
post on our site. Keep up the good writing. https://camillacastro.us/forums/profile.php?id=169911
Hello, I think your site may be having browser compatibility problems.
Whenever I take a loook at youhr sire in Safari, it loos fjne howevfer when opening
inn I.E., it’s got some overlapping issues. I just wanted to prvide you with a quick heads up!
Besides that, fantastic blog! https://depot.lk/user/profile/29827
Hello, I enjoy reading all of yiur post. I like to write a little
comment to supporft you. http://another-ro.com/forum/viewtopic.php?id=152430
Howdy very nice website!! Guy .. Excellent .. Amazing ..
I will bookmark your website and take the feds also? I’m happy to search out numerous helpful information here within tthe
publish, we need work out extra techniques
in this regard, thank you for sharing. . . . . . http://links.musicnotch.com/denicektm213
Appreciation to my father who stated to me regarding this blog,
this blog is genuinely remarkable. http://www.ozsever.com.tr/component/k2/itemlist/user/406295
It’s going to bbe finish of mine day, but before
end I am reading this enormous piece of writing to increase my know-how. https://camillacastro.us/forums/viewtopic.php?id=327072
After checking out a handful of the blog posts on your website,
I seriously like your technique of writing a blog. I book-marked it to my bookmark site
lkst aand will be checking back soon. Take a look at my website too and tell me your opinion. http://another-ro.com/forum/viewtopic.php?id=150728
Hi, its good post on the topiuc off media print, wee all
be familiar with media is a wonderful source of facts. https://camillacastro.us/forums/viewtopic.php?id=327113
Good information. Lucky me I ran across your blog by
accidnt (stumbleupon). I’ve saved itt for later! http://forum.altaycoins.com/viewtopic.php?id=695203
A fascinating discussion iis definitely worth comment.There’s no
doubt that that you ought to publish more on this topic, it may not be a taboo subject but generally folks don’t speak
about such issues. To the next! All the best!! https://www.erotikanzeigen4u.de/author/lazarov8763/
This powt is invaluable. Where can I find out more? http://another-ro.com/forum/viewtopic.php?id=152157
My brother suggested I mighht like this blog. He wass totally right.
This post actually made my day. You cann’t imagine simply how much time
I had spent for this info! Thanks! https://camillacastro.us/forums/viewtopic.php?id=327964
Hello to all, how is everything, I think every one
is getting more from this website, and your views are pleasant in favor of new viewers. http://forum.ainsinet.fr/profile.php?id=351732
Every weekend i used to pay a quick visit this web page, for the rewson that i wish for enjoyment, aas tgis this website cconations in fact pleasant funny dsta too. https://bicycledude.com/forum/profile.php?id=1523896
It’s the best time to make a few plans for the
future and it is time to be happy. I have learn this put up aand if
I mayy I want to recommend you few attention-grabbing issues or
tips. Maybe you can write next articles regarding this article.
I wish to rrad even more things approximately it! http://another-ro.com/forum/viewtopic.php?id=152447
I’m not sure exactly why but this web site is loading incredibly
slow for me. Is anyone else having this issue or iis it a problem on my end?
I’ll check back later onn and see if the problem stijll exists. http://forum.altaycoins.com/viewtopic.php?id=693853
This design is incredible! You mosdt certainly know how to keep
a reader amused. Between your wit and your videos, I was almost moved too start my oown blog (well,
almost…HaHa!) Excellent job. I really enjoyed what you
had to say, and more than that, how you presented it.
Too cool! https://www.fionapremium.com/author/maryannlea/
Touche. Solid arguments. Keep up the great spirit. http://another-ro.com/forum/viewtopic.php?id=152055
Hi, everything is giing fine here and ofcourse every one is sharing data, that’s genuinely excellent, keep up writing. http://kartalescortyeri.com/author/temeka2292/
Hurrah, that’s what I was exploring for, whwt a information! present here at this weblog, thanks admin of this web page. http://kartalescortyeri.com/author/giafrayne7/
Howdy! I could have sworn I’ve been to this website before but after
browsing through many of the articles I realized it’s new to me.
Regardless, I’m certainly pleased I cae across it and I’ll be book-marking it and checking back frequently! http://links.musicnotch.com/aleida79063
I get pleasure from, result in I discovered just what I was having a look for.
You’ve ended my 4 day lengthy hunt! God Bless you man. Have a nice day.
Bye http://another-ro.com/forum/viewtopic.php?id=152103
This article is iin fact a pleasant one it assists new thhe web people, who are wishing in favor of blogging. https://www.alonegocio.net.br/author/arnoldkidwe/
I really like what you guys end to be up too. Thhis sort of clever work and reporting!
Keep up the amazing works guys I’ve added you gujys to my blogroll. http://another-ro.com/forum/viewtopic.php?id=152259
This site really has all of the info I wanted concerning this subject
and didn’t know who to ask. https://camillacastro.us/forums/viewtopic.php?id=328044
Asking questions are actually good thing if you are not understanding anything completely, but this article
offers pleasant understanding yet. http://forum.altaycoins.com/viewtopic.php?id=695942
Awesome blog! Is your theme custom made or ddid you download it from somewhere?
A design like yors with a few simple tweeks would
really make mmy blog stand out. Pleease let me know where you got your theme.
Thanks http://another-ro.com/forum/viewtopic.php?id=152179
Apppreciate the recommendation. Let me try iit out. http://alpervitrin40.xyz/author/melvinaspea/
Pretty! This was a really wonderful post.
Thanks for supplying these details. https://www.fionapremium.com/author/kathysnoddy/
Magnificent website. Plenty of useful info here.
I’m sending it to a few buddies ans also sharing in delicious.
And naturally, thanks in your effort! https://camillacastro.us/forums/viewtopic.php?id=327151
Hello! Do you know if they make aany plugins tto safeguard against hackers?
I’m kinda paranoid about losing everything I’ve worked hard on. Any tips? https://migration-bt4.co.uk/profile.php?id=306250
I don’t even know how I ended up here, but I thought this post was
good. I do nnot know who you are but definitely you’re going to a famous blogger if you
aren’t already 😉 Cheers! https://utahsyardsale.com/author/steffen71i/
I constantly emailed this blog post page to all my friends,
because if like to read it next my links will too. http://forum.altaycoins.com/viewtopic.php?id=696112
Very shortly thios site will be famous amid all blogging and site-building users, ddue to it’s pleasant content https://www.erotikanzeigen4u.de/author/titusb6190/
I loved as much as you will receive carrdied out right here.
The sketch is tasteful, your authored material stylish.
nonetheless, you command get bought an impatience over
that yoou wish bee delivering the following. unwell
unquestionably coome further formerly again since
exactly the samje nearly a lot oftren inside case you shield this increase. http://another-ro.com/forum/viewtopic.php?id=152422
Does your blog have a contact page? I’m having problems locating it but, I’d like to shoot you an email.
I’ve got some recommendations for your blog you might be
interested in hearing. Either way, greatt blog and I look forward to seeing it grow over time. http://forum.altaycoins.com/viewtopic.php?id=693830
I am regular visitor, howw are yoou everybody? Thhis artcle
posted at this web sitge is actually pleasant. http://another-ro.com/forum/profile.php?id=157235
continuously i uxed to read smaller articles which also clear their motive, and that is also happening with this post which I am reading at
this place. http://forum.altaycoins.com/viewtopic.php?id=695325
Hello! Quick question that’s completely off topic. Do you knokw how to
make your site mobile friendly? My weblog loioks weird when browsing
from my iphone4. I’m trying tto find a template or plugin that might be able
to fix this problem. If you have any suggestions, please share.
Many thanks! https://camillacastro.us/forums/viewtopic.php?id=328950
Hurrah! At last I got a web sife from where I be able to genuinely obtain valuable
data regarding my study and knowledge. https://www.fionapremium.com/author/charagranvi/
Hi! Someone in my Myspace group shared ths website with
us so I came to give it a look. I’m definitely enjoying the information. I’m bookmarking
and will be tweeting this to my followers! Excellent bog
aand outstanding design. https://depot.lk/user/profile/29843
Howdy just wanted to give you a quick heads up. The words in your article seem to be running off the screen in Opera.
I’m not sure if this iis a formatting issue orr something to do with browser compatibility but I figured I’d post tto let you know.
The style and design look great though! Hope yyou get the problem resolved soon. Kudos https://camillacastro.us/forums/viewtopic.php?id=327617
What’s up to every one, since I am really eager oof reading thiks
weblog’s post to be updated regularly. It carries pleasant stuff. http://forum.altaycoins.com/viewtopic.php?id=696142
I’m not sure exactoy why but this weblog is loading very slow for
me. Is anyone else having this issue or is it a problem on my end?
I’ll check back latewr on and see if the problem still exists. https://98e.fun/space-uid-7734978.html
Normally I don’t ead post on blogs, but I would like to say that tthis write-up
vrry forced mme to try and do it! Your writing style has been amnazed me.
Thank you, quite great post. http://kartalescortyeri.com/author/navemelia7/
This is my first time ggo to see at here annd i am in fact impressed to read alll at alone place. http://another-ro.com/forum/viewtopic.php?id=151970
What’s up Dear, are you really visiting this website regularly, if so then you will absolutely takoe good know-how. https://www.fionapremium.com/author/lillianaall/
Hey would you mind letting me know which web host you’re working with?
I’ve loaded your blog in 3 completely different internet browsers and I must say this blog
loads a lot quicker then most.Can you recommend a good
internet hosting provider at a fair price? Kudos, I appreciate it! http://another-ro.com/forum/viewtopic.php?id=150527
I am sure this article has touched aall the internet viewers, its really really fastidious piece of writing on building up new website. https://camillacastro.us/forums/profile.php?id=169980
Thanks a bunch for sharing this with all people you really know what
you’re talking about! Bookmarked. Kindly also visit myy web
site =). We may have a hyperlink exchange agreement among us https://oncallescorts.com/author/willisvando/
hey there and thank you for your info – I’ve definitely picked up anything
new from right here. I did however expertise several tchnical issues using this web site, since I experienced
tto reload the site lots of times previous tto I could get
it to load correctly. Ihad been wondering if your web host is OK?
Not that I’m complaining, but sluggish loading instances times will sometimes affect your placement
iin google and can damage your quality scode iif advertising and mareketing with Adwords.
Anyway I am adding this RSS to my email and can look out for a lot moe
of your respective intriguing content. Make sure you update this
again vwry soon. https://depot.lk/user/profile/29819
Hello, I enjoyy reading thurough your article post. I like
to write a little comment to support you. http://another-ro.com/forum/profile.php?id=157183
Hello There. I found your weblog the use of msn. That is a reaslly smartly written article.
I’ll be sure to bookmark it and return to learn extra of
your helpful info. Thanks for the post. I will definitely return. https://migration-bt4.co.uk/profile.php?id=304705
Pretty nice post. I just stumbled upon your blog and
wished to say that I have truly enjoyedd surfing arouhd your
blog posts. After all I will be subscribing to your feed and I hope
yyou write again very soon! https://oncallescorts.com/author/antwanoswal/
Everry weekend i used to go to see this web site, because i wish for enjoyment, aas this this web page conations
actually fastidious funny data too. https://camillacastro.us/forums/viewtopic.php?id=327848
Heya i am for the first time here. I came across this board and I in finding It truly helpful & it helped me oout a lot.
I am hoping to offer something back annd aid others such as you helped me. https://camillacastro.us/forums/viewtopic.php?id=327316
I amm regular reader, how are you everybody? Thiis article posted
aat this web page is actually nice. http://kartalescortyeri.com/author/melissaclar/
Hi there, You have done a fantastic job.
I will definitely digg it and personally recommend to my friends.
I am confident they will be benefited from this website. http://alpervitrin40.xyz/author/margaretaco/
I amm sure this pioece of writing hhas touched all the internet users, its really really pleasant paragraph on building up new webpage. https://migration-bt4.co.uk/profile.php?id=306494
This design is incredible! You most certainly know hoow to keep a reader
entertained. Between your wit and your videos, I was almost moved
to start my own blog (well, almost…HaHa!) Excellent job.
I really loved what you had to say, and more than that, how you presented it.
Too cool! http://www.ozsever.com.tr/component/k2/itemlist/user/406285
It’s remarkable to go to see this site annd reading the views of
all mates on tthe toic of this piece of writing, while I am also keen of getting familiarity. http://forum.altaycoins.com/viewtopic.php?id=695827
Keepp on writing, great job! http://forum.altaycoins.com/viewtopic.php?id=695206
Hi there it’s me, I am also visiting this wweb page regularly, this websitfe is tduly good and the visitors are actually sharing
fastidious thoughts. https://www.mazafakas.com/user/profile/4174856
Everyohe loves it wgen individuals geet together aand
share ideas. Great site, stick with it! https://98e.fun/space-uid-7730730.html
I’m not sure exactly why but this blg iss loading very slow for me.
Is anyone else having this problem or is it a issue on myy end?
I’ll check back later and see if thhe problem still exists. http://links.musicnotch.com/jaquelinepas
Hello There. I found your blog using msn. This is an extremely well written article.
I will make sure to bookmark it and retuhrn to read more of your useful information. Thannks for the post.
I’ll certainly return. https://forum.fne82.org/profile.php?id=492058
I am curious to find out what blog system yyou happen to be
using? I’m experiencing some smaall security issues with
my latest blog andd I’d like to ind something more safeguarded.
Do you have any recommendations? http://forum.ainsinet.fr/profile.php?id=351736
It’s ann awesome article foor all the online people; thsy will obtin benefit from it
I am sure. http://forum.altaycoins.com/viewtopic.php?id=695204
I think that everything said made a great deal of sense. But, what about this?
what iif you were to write a awesome headline? I mean, I don’t wish to tell youu hoow to run your blog,
however suppose you added a headline that grrabbed folk’s attention? I mean Extreme environments
are coded into the genomes of the organisms that live there, research suggests – AI List Line is kinda boring.
You might look at Yahoo’s front page and watch hhow they create news headlines
to grab viewers to open the links. You might add a
related video or a related picture or two to grab people eexcited about everything’ve written. In my opinion, it might bring your posts a little bit
more interesting. http://links.musicnotch.com/lolacorrea14
Write more, thats all I have to say. Literally,
it seems as though you relied on the video to make your point.
You clearly know what youre talking about, why throw away your intelligece on just posting videos to
your site when you could be giving uus something enlightening
to read? http://links.musicnotch.com/darlaguidi38
Hey I know this is off topic but I was wondering if youu knew of any widgets I
could aadd to mmy blog that automatically ttweet
my newest twitter updates. I’ve been looking for a plug-in like this for quite
somje time and was hoping maybe you would have some experience
with something like this. Please let mme know if yoou run into anything.
I truly enjoy reading your blog and I look forward to your new updates. http://www.ozsever.com.tr/component/k2/itemlist/user/406303
Every weekend i used to pay a quick visit this website, for the reason that i wish for
enjoyment, as thnis this web site conations genuinely nice funnyy information too. https://telegra.ph/What-is-the-Difference-between-a-Nonprofit-Organization-and-a-Charity-05-22
Everything is vesry open with a precise description of the issues.
It was truly informative. Your site is very helpful.
Many thanks for sharing! https://664df6c79ffdb.site123.me/
Nice blog here! Allso your website loads up fast!
What host are you using? Can I get your affiliate link tto
your host? I wish myy siote loaded up as fast as yours lol https://4supports.mystrikingly.com/
Thank you, I have recently been searching for information about this subject for a llong time
and yours is the best I have found out till now.
But, what about the bottom line? Are you positive
about the supply? https://newcommunity4.wordpress.com/
Hey There. I found your blog using msn. This is a vvery well written article.
I will make sure to bookmark itt aand come back to learn extra
of your helpful info. Thanks for thee post. I willl certainly comeback. https://beekinds.blogspot.com/2024/05/what-is-charitable-organization.html
I seriously love your site.. Very nice colors &
theme. Did yoou build this site yourself? Please reoly back aas
I’m looking to create mmy own webssite and would love to find out where you got this
from or just what the theme iss called. Thank you! https://gamingera2.wordpress.com/
Wow, amazing weblog structure! Howw lengthy have you ever been blogging for?
you make blogging look easy. The whole lok of your site is
wonderful, as smartly as the content material! https://664e0e8f1b541.site123.me/
Good day! I just want too offer you a huge thumbs up for your great info
you have got here on this post. I am returning to your web site for
more soon. https://gametrendss.mystrikingly.com/
First off I woulld like to say excdllent blog!
I had a quick question tht I’d like to ask if you don’t mind.
I was interested to find out howw you center yourswelf and clear youhr mind betore
writing. I’ve had a tough time clearing my thoughts in getting my ideas
out. I truly do enjoy writing but it just seems like the first 10 to 15 minutes
are generally lost simoly just trying to figure out howw to begin. Any ideas or hints?
Cheers! https://newgametrends.blogspot.com/2024/05/7-huge-gaming-industry-trends-2024-2027.html
you’re in point of fsct a excelolent webmaster.
The website loading pace is amazing. It seems that you’re doing any distinctive trick.
In addition, The contents are masterpiece. you’ve perforrmed a magnificent activity oon this subject! https://telegra.ph/Top-developments-driving-growth-for-video-games-05-22
It’s a pity you don’t have a donaate button! I’d without
a doubt donate to this outstanding blog! I suppose for now i’ll settle for bookmarking
and adding your RSS feed to my Google account.
I look forward tto brand new upodates and will talk about this site
with my Facebook group. Chhat soon! https://telegra.ph/Viewable-History—Five-Most-Watched-Sporting-Events-In-the-World-05-23
Awesome blog! Is your theme custom made or did you download it from somewhere?
A theme like yours with a few simple adjustements would really make my
blog shine. Please let me know where you got youir design. With thanks https://664e1af1a3ef6.site123.me/
What’s up, constantly i ued to check weebsite posts here in the
early hours iin the daylight, as i enjoy to gain knoiwledge
of more andd more. https://sportevents5.wordpress.com/
Wonderful goods from you, man. I’ve take into accout your stuff prior to and you’re simply too fantastic.
I ctually like what you have received right here, certainly like what you are saying
and the waay durig whifh you are saying it.
You’re making itt entertaining and you continue to
take care of to stay it sensible. I can’twait
to learn far more from you. That is actually a tremendous
website. https://664f35960c8f8.site123.me/
I read this post fully regarding the resemblance of newwest and earlier technologies, it’s aweome article. https://worldsports.mystrikingly.com/
hey there and thank you for your information – I have definitely picked up anything new from right here.
I did however expertise a few technical issues using this web site, as I experienced to reload the
web site lots of times previous to I coould get it
to load properly. I had been wondering if your webb hosting is
OK? Not that I am complaining, bbut sluggish loading instances times will very frequently affect your placement in google and can damage
your high-quality score if advertising and marketing with Adwords.
Anyway I am adding this RSS to my email and can look out for a lot more of
your respective intriguig content. Make sure you update this again soon. https://telegra.ph/Viewable-History—Five-Most-Watched-Sporting-Events-In-the-World-05-22
Yes! Finally someone writes aboht casino. https://futuretechs9.wordpress.com/
Outstanding story there. What occurred after? Thanks! https://664f45d9ae475.site123.me/
Appreciate the recommendation. Let me tryy it out. https://ourfutures.mystrikingly.com/
Hello it’s me, I am alwo visiting this site daily, this web page is in fact nice and the users
are realy sharing pleasant thoughts. https://future-techss.blogspot.com/2024/05/future-technology-22-ideas-about-to.html
Hurrah, that’s what I was searching for, what a data!
existing here at this weblog, thanks acmin of thos site. https://newtop5technology.blogspot.com/2024/05/breakthroughs-that-change-our-lives.html
I have been surfing online more tjan three hours as of late, yet
I never discovered any attention-grabbing article like yours.
It is lovely pric enokugh for me. Personally, iff all website owners aand bloggers made just right content material as you did, the web can be a lot more helpful than ever before. https://664f3d9f49be6.site123.me/
Hey! Would you mind if I share your blog with my myspace group?
There’s a lot of folks that I think would really enjoy your content.
Please let me know. Cheers https://travelbugets.wordpress.com/
Fantastic goods from you, man. I’ve understand your stuff
previous to annd you aree just extremely excellent.
I actually like what you have acquired here, really like what you’re sttating aand the way in which you say it.
You make it enjoyable and you still take care off
to keep it wise. I ccan not wait to read far ore from you.
This is actually a great site. https://howtootravel.blogspot.com/2024/05/8-tips-for-visiting-national-parks-on.html
Hey!Do you know if they make any plugins to safeguard against hackers?
I’m kinda paranoid about losing everything I’ve worked hard on. Any
recommendations? https://664f5aad65be3.site123.me/
Hellko there! Do you know if they make any plugins to assist with Search Engine Optimization? I’m trying
to get my blog too rank for some targeted keywords butt I’m not seeing very
good gains.If you know of any please share. Cheers! https://go2trevel.mystrikingly.com/
Wow, marvelous blog layout! How long have you been blogging for?
you made bloggingg look easy. The overall look of your site is
great, let alone thee content! https://mybesttravelsss.blogspot.com/2024/05/nature-and-wildlife-travel-on-budget.html
fantastic issues altogether, you just received a brand new reader.
Whhat would you suggest in regards to your publish
thqt you just ade a few days in the past? Any sure? https://telegra.ph/Top-11-Graphic-Design-Trends-for-2024-05-23
of course like your web-site but you need to test the
spelling on quite a few of your posts. Many of them are rife with spelling problems and
I findd it very bothersome to inform the reality then again I’ll
surely come back again. https://664f652e84874.site123.me/
I think this is amonhg the most significant information ffor me.
And i’m glad reading your article. But sould remark on some general things, The site style is perfect, the articles is really excellent
: D. Good job, cheers https://digital-techss.blogspot.com/2024/05/10-inspiring-graphic-design-trends-for.html
You are so interesting! I don’t suppose I’ve truly read through
something like that before. So ggood to find someone with a few original thoyghts on this subject matter.
Seriously.. thanks for starting this up. This site
iss something that’s needed on the internet, someone with a little originality! https://newprojectss.mystrikingly.com/
Oh my goodness! Amazinng article dude! Thank you so much,
However I am encountering prolems with your RSS. I don’t understand the reason why I cannot join it.
Is there anybody ese having similar RSS issues?
Anyone who knows thhe solution will you kindly respond?
Thanx!! https://graphicdesign058.wordpress.com/
This information is worth everyone’s attention. When can I find out more? https://caramellaapp.com/milanmu1/Q22q6JTpX/the-history-of-bitcoin-the-first-cryptocurrency
It’s going to be finish of mine day, but before finbish I amm reading this
impressive paragraph to increase my knowledge. https://posteezy.com/what-are-smart-contracts-blockchain-and-how-do-they-work-0
Greetings! Very useful advice in this particular article!
It is the little changes that produce the greatest changes.
Many thanks ffor sharing! https://www.pearltrees.com/alexx22x/item598954865
Heyya i am for thee first time here. I found this board and I find
It really helpful & it helped me out a lot. I am hoping to present one thing agin and aid others such as you aided me. https://www.pearltrees.com/alexx22x/item598913531
Thanks for ones marvelous posting! I really enjopyed reeading it,
yoou might be a great author. I will always bookmark your blog annd will come back in the foreseeable future.
I want to encourage continue your great posts, haave a nice day! https://www.pearltrees.com/alexx22x/item598958345
Howdy! I could have sworn I’ve been to this website before but after cuecking through somje of the post I realized it’s new to
me. Anyways, I’m definitely delightewd I found it and I’ll be book-marking and checkijg back often! https://caramellaapp.com/milanmu1/qApsvUpeV/newtech
Hello to all, the contents existing at this web site are genuinely remarkable for people knowledge,
well, keep up the good work fellows. https://scrapbox.io/digitals/Difference_between_nonprofits,_NGOs_and_charities
Woow that was strange. I just wrote an extremely long comment but after I clicked submit
my cmment didn’t show up. Grrrr… well I’m nott writing all thatt over again.
Anyhow, just wanted to say superb blog! https://caramellaapp.com/milanmu1/idzO9jSXi/top-tech
My relatives every tiome ssay that I am wasting my time here
at net, however I know I am getting knowledge every day by reading such nice articles. https://scrapbox.io/toptech/Top_10_Latest_Technological_Innovations
I costantly emailed this weblog post page to all my associates, since if like too read it after that
my links will too. https://iitmimmigration.com/%d0%be%d0%b3%d0%bb%d1%8f%d0%b4-%d0%ba%d0%b0%d0%b7%d0%b8%d0%bd%d0%be-%d0%b1%d0%b5%d1%82%d0%bc%d0%b0%d1%82%d1%87-%d1%83%d0%ba%d1%80%d0%b0%d1%97%d0%bd%d0%b0/
I have been surfing on-line greater than threde hours today, but
I nevr discovered any interesting article like yours.
It’s pretty price enough for me. In my opinion, if all site owners and bloggers made good content as you probably did, thee internet will bee
a lot more useful than ever before. https://www.pitstopodium.com/f1-news/%d1%80%d0%b5%d1%94%d1%81%d1%82%d1%80%d0%b0%d1%86%d1%96%d1%8f-%d0%b2-%d0%b1%d0%b5%d1%82-%d0%bc%d0%b0%d1%82%d1%87/
I seldom create comments, but I read a few of thee comments on Extreme envieonments arre coded into
thhe genomes of the organisms that live there,
research suggbests – AI List Line.
I actually do hazve 2 questions for you if it’s
okay. Could it be just me oor does it appear like some of
the remarks lpok like they are written by brain dead folks?
😛 And, if you are posting on additional sites, I would
like to keep upp wiyh you. Could you list
of the compete urls of your shared sites like your Facebook page, twitter feed, or linkedin profile? https://aulacorporacionnovasur.cl/blog/index.php?entryid=2474
I’m not that much of a internet reader to be honest but your blogs really nice, keep it up!
I’ll go ahead and bookmark your site to come back in the future.
All the best https://camillacastro.us/forums/viewtopic.php?id=341367
Thanks for ones marvelous posting! I definitely enjoyed
reading it, you might be a great author.I will make certain to bookmark your blog aand wilol come back downn the road.
I want to encourage you continue yur great work,
have a nice day! https://marriagesofa.com/profile/qazfatima14363/
Hello there! Do you know if they make any plugins to protect against hackers?
I’m kinda paranoid about losing everything I’ve worked hard
on. Any suggestions? http://forum.altaycoins.com/viewtopic.php?id=707209
Hello there, You’ve donme ann excellent job.
I’ll certainly digg it and personally recommend tto my friends.
I’m confident they’ll be benefited from this weeb site. https://advansbum.by/?option=com_k2&view=itemlist&task=user&id=889685
Fine way of explaining, and peasant post to obtain data on the topic of my presentation topic, which
i am going to deliver iin institution of higher education. https://marriagesofa.com/profile/scarlett91d7481/
I like the helpful information you supply in your articles.
I will bookmark your blog aand tame a look at onc more here regularly.
I am quite sure I’ll learn plenty of new stuff proper here!
Good luck for the following! https://katazaraki.com/%D0%BA%D0%B0%D0%BA-%D0%B7%D0%B0%D1%80%D0%B5%D0%B3%D0%B8%D1%81%D1%82%D1%80%D0%B8%D1%80%D0%BE%D0%B2%D0%B0%D1%82%D1%8C%D1%81%D1%8F-%D0%BD%D0%B0-bet-match/
Very good article. I am facing some of these issues as
well.. http://kartalescortyeri.com/author/mayrabutche/
My brother recommended I might like this website. He was totally right.
This post actually made my day. You cann’t imagine just
how much time I had spent for this information! Thanks! https://www.mazafakas.com/user/profile/4205430
Way cool! Somme extremely valid points! I appreciate you penning this post and also the rest of tthe website
is really good. https://marriagesofa.com/profile/sabinedumaresq/
I was very happy to find this site. I need to to thank
you for your time just for this fantastic read!! I definitely realy liked
every part of it and I have you book marked to look at new things oon your web site. http://forum.altaycoins.com/viewtopic.php?pid=807272
Hello there, I discovered your web site by the use oof Google even as lookinmg for a comparable subject, you website came up, it seesms
good. I have bopkmarked itt in my google bookmarks.
Hello there, just turned into aware of youur weblog via Google,
and located that it is truly informative. I’m going to watch out ffor brussels.
I will appreciate if you proceedd this in future. Lots of otther people will likely
be benefited out of your writing. Cheers! https://oceancitygames.co.uk/atlantisclubforum/profile/jillwolfgang15
Simply wish to sayy your article is as amazing.
The clearness in your post is just nice and i could
assuume you aare an expert oon this subject.
Well with your permkssion allow me to grab your RSS feed to
keep updated with fothcoming post. Thanks a million and please carry on the
enjoyable work. https://ingeconvirtual.com/bet-match-%d0%b1%d0%be%d0%bd%d1%83%d1%81%d0%b8-%d0%b2%d1%96%d0%b4-%d0%be%d0%bd%d0%bb%d0%b0%d0%b9%d0%bd-%d0%ba%d0%b0%d0%b7%d0%b8%d0%bd%d0%be/
Hi there! This is myy first visit to your blog!
We are a team of volunteers and starting a new iniktiative
in a community in the same niche. Your blog provided us beneficial information to work on. You
have done a extraordinary job! https://institutogdali.com/blog/index.php?entryid=221261
Fantastic goods from you, man. I’ve understand your stuff previous to and
you arre just too wonderful. I really like what you’ve acquired here, certainly lime what you aree
statinhg and the way in which you say it. You make it entertaining and you still take care of to keep it smart.
I can not wait to read far more frolm you. This is actually a great
site. https://careked.com/community/profile/kristiestagg270/
Hey there superb website! Does running a bloog similar to this take a lot of work?
I have virtually no expertise in programming but I was
hoping to start mmy own blog soon. Anyway, if you have any ideas orr tips
foor new blogg owner please share. I know this is off topic but I juxt
wanted to ask. Many thanks! http://forum.altaycoins.com/viewtopic.php?pid=807336