
[ad_1]
T. thermophilus HB27 strain expresses a Cfr-like methylase. Credit: Nature Chemical Biology (2024). DOI: 10.1038/s41589-023-01525-w
Scientists at the University of Illinois Chicago and Harvard University have developed an antibiotic that could give medicine a new weapon to fight drug-resistant bacteria and the diseases they cause.
antibiotic, cresomycin, described I scienceEffectively suppresses pathogenic bacteria that have become resistant to many commonly prescribed antibiotics Antimicrobial drugs.
The promising novel antibiotic is the latest discovery in a long-standing research partnership between the group of Yuri Polykanov, associate professor of biological sciences at UIC, and colleagues at Harvard. UIC scientists provide important insights into this. Cellular mechanisms and structure that help Harvard researchers design and synthesize new drugs.
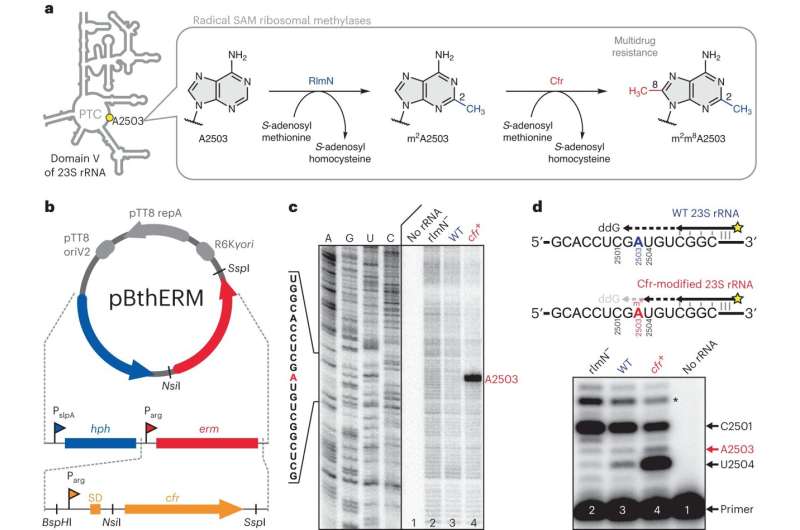
In the development of A new antibioticthe group focused on how many antibiotics interact with a common cellular target. The ribosome-And how? Drug-resistant bacteria Modify their ribosomes to defend themselves.
Inhibits the growth of more than half of all antibiotics Pathogenic bacteria By interfering with their protein biosynthesis – a complex process catalyzed by the ribosome, which is “like a 3D printer that makes all the proteins in the cell,” Polikanov said. Antibiotics bind to the bacterial ribosome and disrupt the production of this protein, causing the bacterial invaders to die.
But many species of bacteria have developed simple defenses against this attack. In a defense, they interfere with antibiotic activity by adding a Methyl group of one carbon and three hydrogen atoms in their ribosomes.
The scientists hypothesized that this defense is simply the bacteria physically blocking the site where the drugs bind to the ribosome, “like putting a push pin on a chair,” Polikanov said. But the researchers found a more complicated story, as they described In a paper published last month Nature Chemical Biology.
Using a method called X-ray crystallography to visualize drug-resistant ribosomes with near-atomic precision, they discovered two defense mechanisms. They found that the methyl group blocks the physical binding sitebut it also changes the shape of the inner “gut” of the ribosome, which further affects the activity of the antibiotic.
Polikanov’s laboratory then used X-ray crystallography to determine that certain drugs, including a published I The nature In 2021, a UIC/Harvard collaboration, Prevent this common form of bacterial resistance.
“By determining the actual structures of antibiotics that interact with two types of drug-resistant ribosomes, we saw what could not have been predicted by available structural data or computer modeling,” Polikanov said. ” “It’s always better to see it once than hear about it 1,000 times, and our structures were critical to designing this promising new antibiotic and understanding how it avoids the most common forms of resistance. can.”
Cresomycin, the new antibiotic, is synthetic. It is prearranged to avoid the interference of the methyl group and binds tightly to ribosomes, disrupting their function. The process involves locking the drug into a form that is already suitable for binding to ribosomes, which helps it get around bacterial defenses.
“It just binds to the ribosome and acts like it doesn’t care whether it was methylated or not,” Polikanov said. “It easily overcomes many common forms of drug resistance.”
I Animal experiments Developed at Harvard, the drug protects against infections with multidrug-resistant strains of common disease drivers including Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa. Based on these promising results, the next step is to evaluate the efficacy and safety of cresomycin in humans.
But even at this early stage, the process demonstrates the important role that structural biology plays in the design of the next generation of antibiotics and other life-saving drugs, according to Polikanov.
“Without the structure, we would be blind to how these drugs bind to and act on drug-resistant ribosomes,” Polikanov said. “The structure we determined provided fundamental insight into the molecular mechanisms that allow these drugs to evade resistance.”
In addition to Polikanov, UIC co-authors include Elena Alexandrova, Igor Serugin and Maxim Svetlov. science Paper and Aleksandrova, Syroegin, Svetlov and Samson Balasanyants on the Nature Chemical Biology Paper
More information:
Elena V. Alexandrova et al., Structural basis of Cfr-mediated antimicrobial resistance and mechanisms of its avoidance, Nature Chemical Biology (2024). DOI: 10.1038/s41589-023-01525-w
Provided by
University of Illinois at Chicago
Reference: New research helps create new antibiotic that evades bacterial resistance (2024, Feb 17) Accessed 17 Feb 2024 at https://phys.org/news/2024-02-antibiotic-evades-bacterial-resistance.html Obtained from
This document is subject to copyright. No part may be reproduced without written permission, except for any fair dealing for the purpose of private study or research. The content is provided for informational purposes only.
[ad_2]


